Improving Axon Tracing and Centerline Detection to Study the Brain
An adaptation combining two deep learning techniques has been shown to provide a successful automated process for axon tracing and centerline detection in brain microscopy images. Axon tracers are powerful tools providing crucial insight into the development, connectivity, and function of neural pathways. Automating this process alleviates personnel time and costs involved in manual tracing. Scientists, equipped with advanced algorithms and processing, will soon be able to develop insights from high-resolution, networked brain atlases that illuminate underlying differences between healthy and diseased brains.
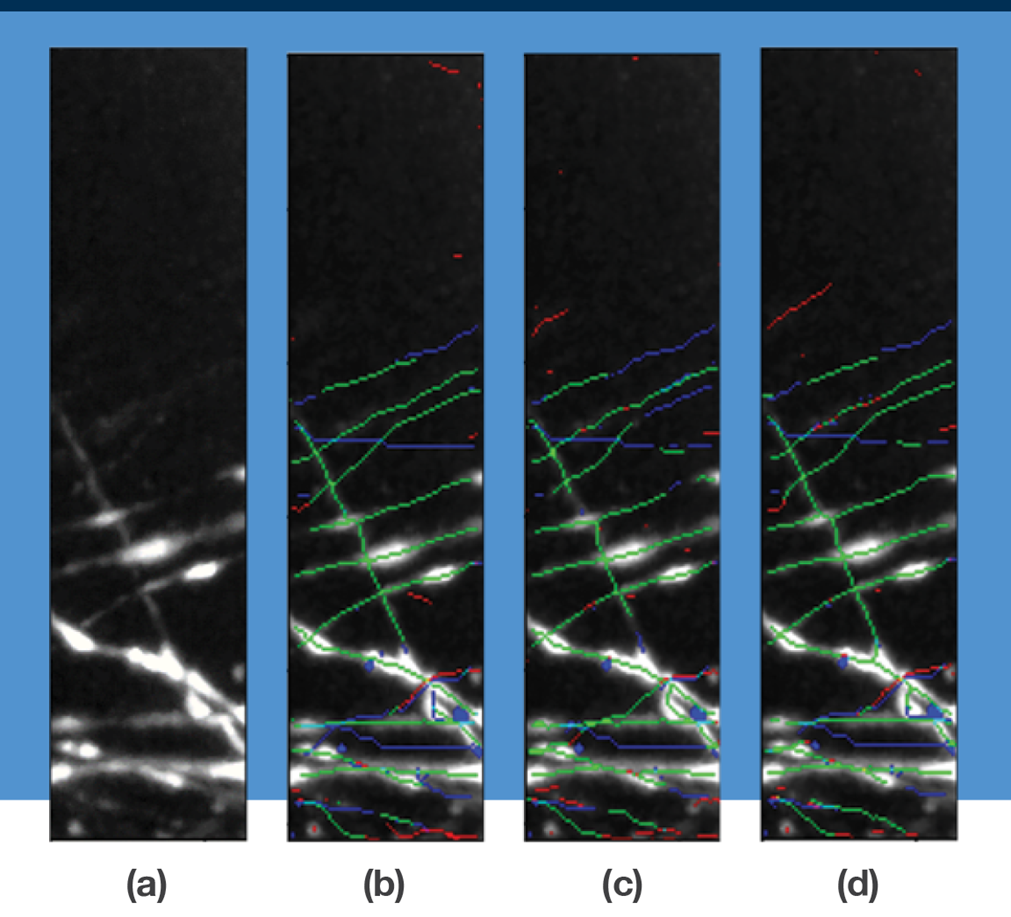
The figure illustrates results from experiments to trace axon centerlines using various neural network tools. In (a) is a 3D microscopy image of mouse brain tissue showing the maximum intensity projection along the z-axis. In (b), the tracing is generated via 3D U-Net; in (c) via CasNet; and in (d) via 3D U-Net + clDice. The tracing results are overlaid with true positive centerline predictions (green), false negatives (blue) and false positives (red). 3D U-Net + clDice results show fewer fragmenting errors caused by false-negative predictions.
Motivation
Improved high-resolution brain imaging capabilities have yielded a trove of data useful for tracing neural connectivity and correlating it with brain functions. However, human manual tracing is a slow, laborious task that requires domain expertise. Automated methods enabling rapid and accurate analysis of the billions of connections would expand research into the complex interactions of human brain.
Toward a Better Method
Researchers from Lincoln Laboratory teamed with the Chung Lab at MIT to investigate automated methods to generate axon tracings and predict axon centerlines that ground the tracings. This work aids the collective efforts of researchers in transcriptomics, neuroanatomy, neurophysiology, computational biology, and microscopy to accelerate progress in decoding brain function. The Chung Lab has made human brain tissue data available at https://dandiarchive.org/dandiset/000108.
The team conducted experiments evaluating the performance of three convolutional neural networks in segmenting neuron data and predicting an axon's centerline. The experiments were done using a dataset of light-sheet microscopy of mouse brain tissue. A residual 3D U-Net architecture was employed to carry out classification tasks; CasNet, a cascading network previously used in road detection, was used for segmentation and centerline detection; and a soft-centerline Dice (clDice) metric that incorporates parameter-free skeletonization (i.e., reduction of an object to its key outline) was used to evaluate a generated centerline vs the actual one.
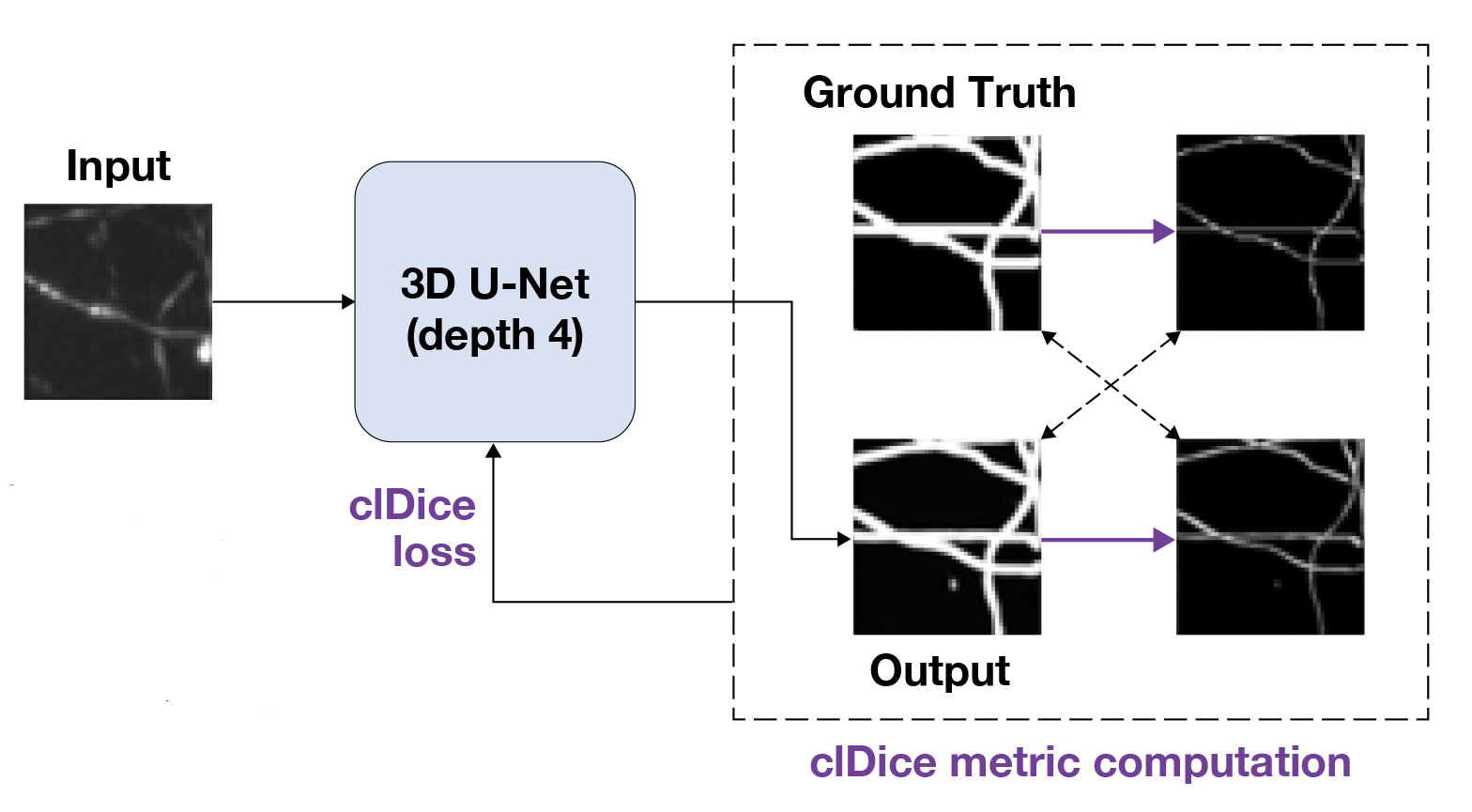
The Soft-clDice approach thins the 3D U-Net segmentation output using soft-skeletonization (purple arrows) so that centerline information can be incorporated into the loss function.
A hybrid making use of the 3D U-Net and soft-clDice tools had very good performance in both centerline prediction and axon tracing. This technique shows potential for improved feature extraction for large-scale brain mapping. Further research examining the use of 3D U-Net + soft-clDice with other machine learning approaches, including self-supervised learning, could refine the tool for wider use.
Benefits
- Accuracy of axon tracing increased by 10% over previous methods
- Tracings from the combined method show fewer splitting errors caused by false-negative predictions
- The technique has potential for further refinement to improve centerline detection
- The method improves connectivity of long-range fibers
- An automated starting point enables validation of dense neuronal regions
Additional Resources
This software is available at https://github.com/mit-ll/axon-centerline-detection
Publications
D. Pollack, L.A. Gjesteby, et al., "Axon Tracing and Centerline Detection Using Topologically-Aware 3D U-Nets," in 44th Annual International Conference of IEEE Engineering in Medicine & Biology Society, 11−15 July 2022.
J. Park et al., "Integrated Platform for Multiscale Molecular Imaging and Phenotyping of the Human Brain," Science, vol. 384, no. 6701, 14 June 2024. The human brain tissue data used in this paper is available at https://dandiarchive.org/dandiset/000108